Sensing and processing chemical signals.
We investigate how individual cells and cell populations detect the intensity and identity of chemical signals, and the role of fluctuations in this process.
Chemical sensing in bacteria. We have showed that the two-component system E. coli bacteria use to detect and process chemical signals for navigation is very noisy. These results, recently confirmed by direct single-cell FRET measurements of the kinase activity, raise fundamental questions about chemical sensing. Are fluctuations just noise, or do cells use them to compute? How do cell growth and division affect signal processing? Are the distributed signal-processing abilities of an isogenic population of bacteria more than the sum of those of its constituents? To tackle these questions, we combined our microfluidics technology with the single-cell FRET system from collaborator Tom Shimizu to measure response variability within and across cells simultaneously (Fig 1).
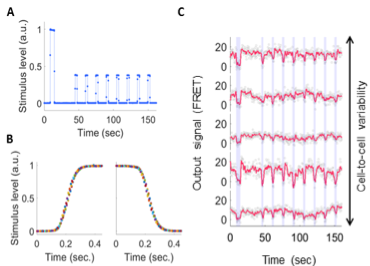
Figure 1. Single-cell FRET measurement in a microfluidic device. A. Input stimulus. B. Stimulus can be switched in 0.1 seconds (colors are repeated pulses). C. Single-cell FRET response to methyl-aspartate (dots). Red: smoothed over 1.5 s. Blue shades: time points where stimulus is high.
From individuals to emerging group composition, structure, and function
In biology, functions are typically carried by groups of cells that express similar genes, yet exhibit phenotypic diversity. A central focus of our lab is to discover basic rules by which phenotypic diversity modulates isogenic populations’ performance, and how function and spatial organization at the scale of the group (microbial population, developing tissue) emerge from interactions and coordinated behavior of individual cells.
By racing isogenic bacteria in a static gradient of attractant, we discovered that phenotypic diversity can cause populations to outcompete their mean phenotype. This happens because the mappings from protein abundance to phenotype to performance are nonlinear. Thus, the population-average performance is different from the performance of the average individual. Next, we examined how phenotypic diversity and collective behavior affect each other when cells migrate collectively by consuming an attractant in the environment, which generates a traveling gradient that they chase. We discovered that bacteria spontaneously sort themselves along the traveling gradient, resolving the conflict between cells’ different individual performances and traveling together as a group (Fig. 2). Thus, Julius Adler’s classic waves are not well-mixed: a leader-follower organization of the phenotypes emerges, even in isogenic populations.
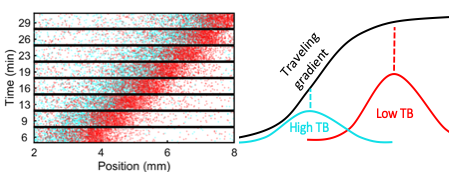 Figure 2. Spontaneous spatial organization of phenotypes during collective migration resolves the conflict between individuality and collective migration. A one-to-one mixture of E. coli cells with high (cyan) and low (red) tumble bias (TB) spontaneously sort themselves during travel so that the lower tumble bias cells, which climb gradient faster are in the front where the gradient is shallower and the high TB cells, which are slower, are in the back where the gradient is steeper
Figure 2. Spontaneous spatial organization of phenotypes during collective migration resolves the conflict between individuality and collective migration. A one-to-one mixture of E. coli cells with high (cyan) and low (red) tumble bias (TB) spontaneously sort themselves during travel so that the lower tumble bias cells, which climb gradient faster are in the front where the gradient is shallower and the high TB cells, which are slower, are in the back where the gradient is steeper
Integrating sensation and behavior to navigate.
We investigate how organisms integrate new information measured by their sensors with past evidence to make decisions and navigate. Central to this problem is the realization that past decisions influence the stimuli experienced next.
Run-and-tumble navigation. During run-and-tumble navigation used to climb gradients, we found that the feedback of a cell’s motion onto the signals it experiences profoundly affects its performance and can cause it to ratchet up gradients much faster than previous theories predicted. With collaborator Ben Machta from Physics, we are examining the impact of this behavioral feedback on information processing. By measuring drift speeds and signal transduction responses and fluctuations in single cells, we are determining how efficiently E. coli uses information about the gradient during chemotaxis. Our goal is to gain a comprehensive mechanistic understanding of signal processing and navigation at the single-cell level.