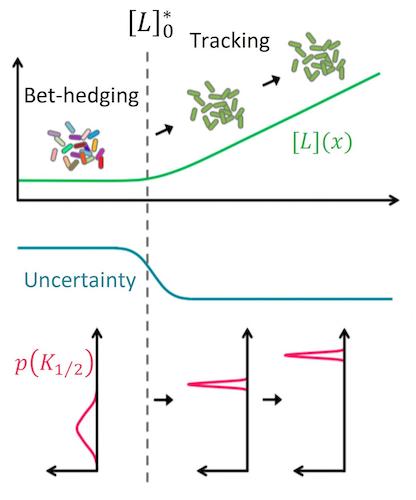
Keita’s paper in Science Advances is out: he discovered a mechanism that allows an isogenic population of bacteria to rapidly adjust its phenotypic diversity when uncertainty in the environment changes. When there is no signal, uncertainty about what signal may come next is high and the population edges its bets by exhibiting strong diversity in the sensitivity of the individual cells (broad distribution in K1/2) to many different signals. But once a given signal L exceeded a threshold (L0 predicted by the mathematical model), the sensory diversity collapsed, enabling the entire population to ‘focus’ on that particular signal. The mechanism mediating this diversity tuning combines allosteric interactions and covalent modifications between protein molecules. Thus, it can take place much faster than known mechanisms that involve gene expression. Such protein modifications are used widely to process and relay signals inside individual cells, but it was not previously known that they could also play a role in the diversification of entire cell populations. Because such modifications are so prevalent throughout cells of all organisms, this basic mechanism for fast changes in cellular diversity could be at work in many cell types across biology. This is a collaboration with Tom Shimizu’s lab at AMOLF. The work was started there and finsihed here.
Read the paper (html, pdf, suppinfo) and a nice summary in EurekaAlert: Dynamic risk management in cell populations